Murn Lab
For Creative Research of RNA Biology
INSPIRATION
The Murn lab draws inspiration from the central role that RNA as life's indispensable molecule plays in biology. We are committed to solving fundamental problems in the field of RNA biology to illuminate life at the level of molecules, cells, and organisms while keeping a keen eye on potential practical implications. We are especially interested in how RNA-binding proteins (RBPs) regulate various events during the lifetime of an RNA molecule to control gene expression and thereby direct the workings of a cell. For instance, we investigate how RNA processing by RBPs regulates cellular quiescence, how it supports the functioning of neurons, and how its misregulation contributes to aging and human diseases, including cancer and neurological disorders. We are inspired by the prospect that our discoveries may contribute to development of new therapies for these diseases.
Our studies make heavy use of genome-wide and computational approaches in combination with traditional biochemistry, genetics, and cell biology to allow for systems-level understanding of RNA networks and their regulatory principles. We also develop novel technologies to decipher in vivo protein-RNA and RNA-RNA interactions and their functions.
Some of our recent stories
-
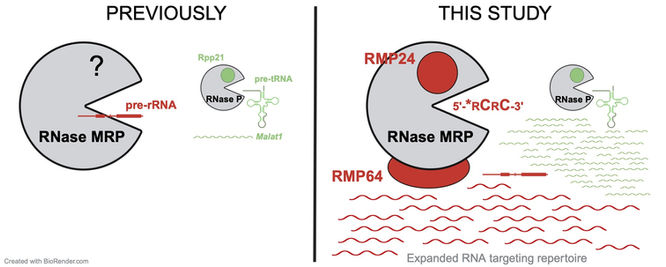
Composition and RNA binding specificity of metazoan RNase MRP
Maturation of rRNA in eukaryotic cells requires cleavage by a half-megadalton ribonucleoprotein, RNase MRP. A vexing question emerges: why is such a large, RNA-based complex required for a simple hydrolytic cleavage of one RNA substrate? In this paper, we show how specific protein subunits distinguish RNase MRP from related enzymes and we unravel its RNA targeting repertoire in mammalian cells. Our study identifies two constitutive protein subunits, which we name RMP24 and RMP64, that are unique to metazoan RNase MRP and absent from the closely related RNase P, which processes tRNA precursors. We leverage our discovery to identify an unexpectedly broad suite of candidate RNA substrates of each enzyme in mammalian cells, including positions of their cleavage at nucleotide resolution. Find our study at Nucleic Acids Research.
-
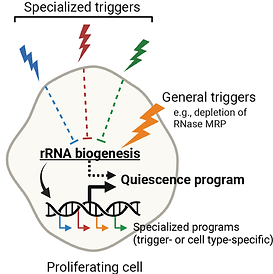
Cellular quiescence induced by rapid depletion of RNase MRP
Most cells in the biosphere reside in quiescence – a state of reversible proliferative arrest – which can last from a few days to several decades. Quiescence plays critical roles in development, resistance to stress, reproduction, tissue repair, immunity, aging, and longevity of organisms. How cells enter quiescence and adapt their homeostasis to a long-lasting nondividing state is an important unsolved problem in biology. In this study, we report a striking finding that rapid depletion of RNase MRP, an RNA-based enzyme that is required for rRNA biogenesis in eukaryotes, induces quiescence of human cells. The cells arrest at all stages of the cell cycle, remain viable for at least several weeks, and can resume proliferation at the speed of control cells upon restoration of RNase MRP. Read more in our paper at Nature Communications.

-
A pseudouridine-binding protein


Pseudouridine (Ψ) is the most abundant RNA modification in nature; however, not much is known about the biological functions of this modified nucleoside. In collaboration with Yinsheng Wang's lab, we employed quantitative proteomics and iCLIP to identify a cytoskeletal protein profilin-1 (PFN1) as a reader of Ψ in cellular RNA. This study also demonstrates that PFN1—Ψ interactions are crucial for regulating the stability and translation efficiency of mRNAs. Check out our paper published in the Journal of the American Chemical Society.
-
A paradigm for regulation at the effector interface with RNA-binding proteins
How RBPs convey regulatory instructions to the core effectors of RNA processing is unclear. We solved this puzzle for Unkempt by dissecting its effector interface. We identified an array of linear motifs that support direct contact with two central effectors of Unkempt, the CCR4-NOT complex and PABPC. We found that the spatial arrangement, dual-purpose motifs, and obligate homodimerization are required for high-avidity effector interactions that are mandatory for the accuracy of RNA targeting, translational repression, and control of cell morphology by Unkempt. We establish the molecular assembly and functional principles of an RBP–effector interface, with general implications for the evolution and function of RBP-operated regulatory networks. Find the story in Nature Communications.

-
The nexus between RNA-binding proteins and their effectors
Beyond their recognition of RNA and their general functions, little is still known about how RNA-binding proteins (RBPs) mechanistically regulate RNA processing. Our analysis shows that numerous RBPs converge onto fewer effectors of RNA processing, forming a distinct regulatory level that contributes to the hierarchical organization of RNA networks. We find that RBP–effector interactions are dominated by contacts between short linear motifs within intrinsically disordered regions of RBPs and structured domains of effectors, allowing for high specificity but transient nature of the interactions. The RBP–effector interface serves as a versatile platform that senses intra- or extracellular stimuli and converts them into biological responses via transient or stable adjustment of RNA processing. We further find that dysregulation of contacts between RBPs and their effectors often causes human disease and that pharmacological or genetic targeting of the RBP–effector nexus carries significant therapeutic potential for neuromuscular disorders, immune disorders, and cancer. Find our story in Nature Reviews Genetics.

-
Post-translational modulation of RNA-binding proteins

Reversible regulation of RBP–effector contacts is typically achieved by PTMs, of which phosphorylation has arguably garnered the most attention, particularly in the context of signal-regulated RNA processing events. The fast turnover of phosphorylation in fact renders this PTM particularly well-suited to mediating rapid responses of several types of RNA processing to a variety of signals. We found that the activity of the RNA-binding protein Unkempt, a key regulator of cell morphogenesis, is regulated by nutrient levels and growth factors via phosphorylation by mTORC1. Find out more about this story at Journal of Biological Chemistry. .

-
A closer look identifies novel small regulatory RNAs
High-throughput RNA sequencing (RNA-seq) has greatly advanced small non-coding RNA discovery, but the widely used library construction protocols often give rise to biased sequencing results. We collaborated with Qi Chen's group to develop an improved library preparation method, called PANDORA-seq, that identified numerous previously unseen small regulatory RNAs primarily derived from tRNAs and rRNAs. Read the story at Nature Cell Biology.

-
Control of cell morphogenesis by an RNA-binding protein

Cell shape is one of the most distinctive features of all somatic cells yet little is known about how it is specified. We discovered a molecular program, controlled at the RNA level, that determines the early shape of neurons. We found that a deeply conserved RNA-binding protein, called Unkempt, coordinates an entire morphology program to establish the early bipolar shape of neurons during embryonic development of an organism. A testament to the power of post-transcriptional control and the activities of RBPs! See the story in Genes & Development.

-
A 'heavy-duty' mode of RNA sequence recognition
Following up on our initial story on the RNA-binding protein Unkempt, we wondered how the six CCCH zinc fingers of Unkempt - the largest such array of any mammalian protein - operate to recognize a relatively short stretch of RNA sequence. We found that the six zinc fingers form two bulky RNA-binding folds, each recognizing a distinct trinucleotide RNA motif. Presence of both motifs and their relative position within coding regions of targeted genes is critical for high-affinity RNA binding, which, in turn, is required for translational repression of Unkempt-target genes. Find the story at Nature Structural and Molecular Biology..

